Agricultural AI: Flower Density Estimation
Advanced Computer Vision for Precision Orchard Yield Prediction
Project Gallery
Image Placeholder 1
Add your project image here

Image Placeholder 2
Add your project image here

Image Placeholder 3
Add your project image here

Image Placeholder 4
Add your project image here
Project Overview
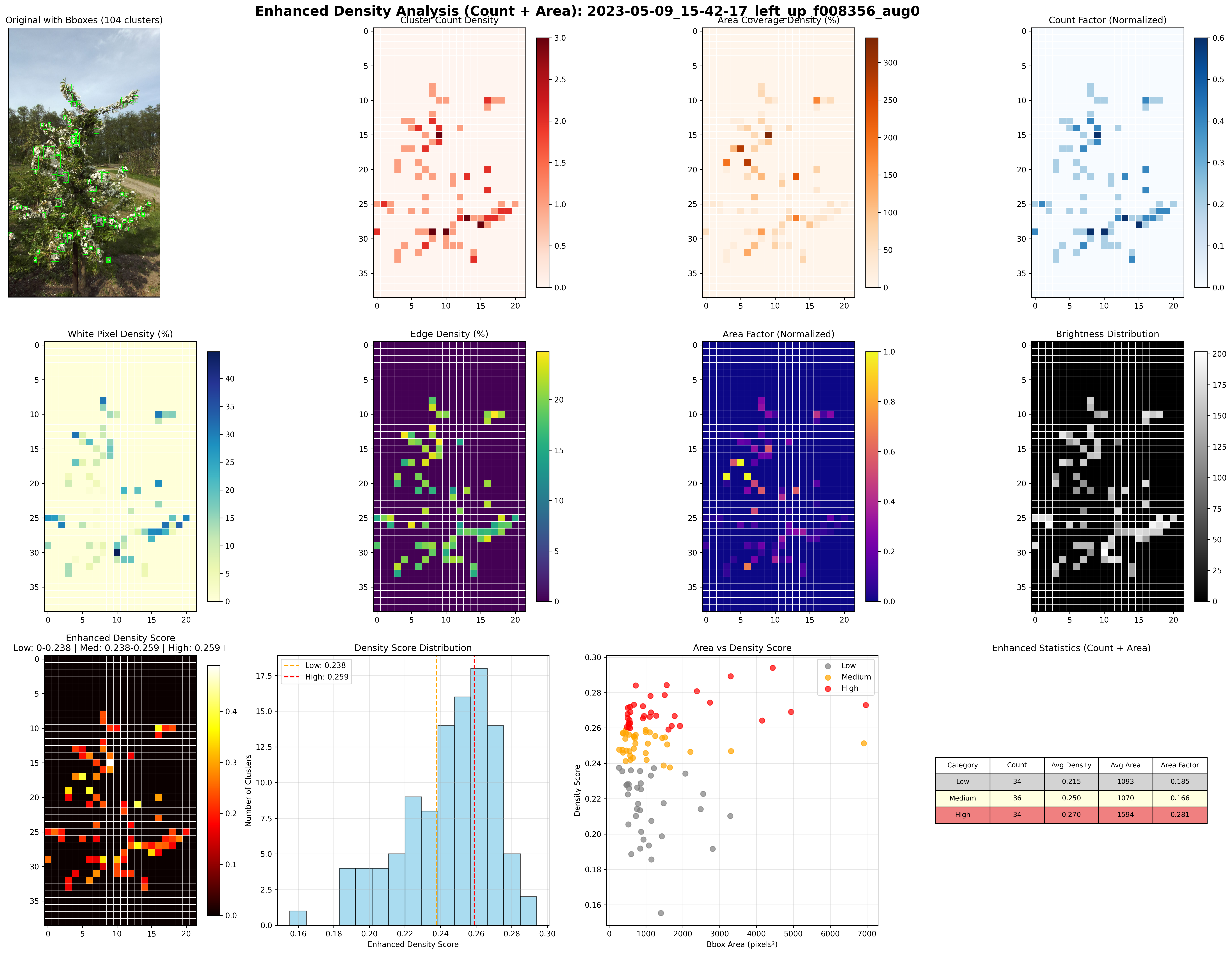
An advanced agricultural computer vision solution that quantifies flower density in tree fruit orchards using deep learning and image analysis. This system combines depth-based background removal, YOLO segmentation, intelligent clustering algorithms, and multi-factor density calculations to provide accurate bloom density assessments critical for yield prediction, crop-load management, and thinning decisions in commercial orchards.
The pipeline integrates Depth-Anything-V2 for monocular depth estimation, custom-trained YOLO models for instance segmentation, DBSCAN spatial clustering for biologically meaningful grouping, and sophisticated density metrics combining flower pixel coverage with edge detection analysis.
Problem Statement
Accurate flower density estimation is essential for orchard management decisions including yield forecasting, optimal thinning, and crop-load planning. Manual counting is labor-intensive and impractical for commercial operations, while flowers present unique detection challenges due to dense clustering, high occlusions, variable lighting, and similar appearance to background foliage. Traditional object detection approaches struggle with densely packed flowers (>100 per image), exhibiting error rates five times higher than regression-based methods in high-density scenarios.
Technical Architecture
Depth Estimation Module
Depth-Anything-V2 generates monocular depth maps for intelligent background separation, isolating foreground flowers from distant foliage
Detection & Segmentation
Custom-trained YOLO model performs instance segmentation with precise polygon masks for individual flowers at pixel level
Clustering Engine
DBSCAN-based spatial clustering with geometric union operations groups flower detections into biologically meaningful clusters
Density Analysis
Multi-factor density calculation combining flower pixel coverage and edge pixel metrics with configurable weighting schemes
Deep Learning & CV Stack
- ▸PyTorch: Core deep learning framework for model inference and tensor operations
- ▸Ultralytics YOLO: Instance segmentation with polygon mask extraction at 640×640 resolution
- ▸Depth-Anything-V2: Vision Transformer (ViT-Large 256 features) for monocular depth estimation
- ▸OpenCV: Image preprocessing, Canny edge detection, color space conversions (RGB/HSV/LAB)
- ▸Scikit-learn (DBSCAN): Density-based spatial clustering with adaptive epsilon tuning
Geometric & Data Processing
- ▸Shapely: Computational geometry for polygon union, intersection, buffer operations
- ▸NumPy: Numerical computing for array operations, quantile calculations, vectorization
- ▸Pandas: DataFrame operations for structured data management and analysis
- ▸OpenPyXL: Excel file generation for comprehensive per-cluster and per-image reports
- ▸Matplotlib & PIL: Visualization and image loading/manipulation capabilities
Core Features & Capabilities
Intelligent Background Removal
Depth-based segmentation leveraging geometric cues to separate foreground flowers from complex backgrounds with 50th percentile threshold
High-Density Flower Detection
Segmentation + regression pipeline maintains accuracy with >100 flowers per image, handling extreme occlusions in full-bloom conditions
Biologically Meaningful Clustering
DBSCAN spatial clustering with adaptive epsilon tuning (0.15-0.30) mirrors natural branch structure with ~8 clusters per image
Configurable Density Metrics
Dual-factor calculation: (flower_weight × Flower%) + (edge_weight × Edge%) customizable for different crop morphologies
Robust Lighting Adaptation
Dynamic brightness adjustment in HSV color space ensures consistent detection from harsh midday sun to overcast skies and shadowed areas
Comprehensive Reporting
Automated Excel generation with per-cluster analysis, image-level summaries, and dataset-wide statistics with visual annotation overlays
Performance Specifications
Key Algorithms & Techniques
🔍 Monocular Depth Estimation (Depth-Anything-V2)
Vision Transformer encoder generates relative depth maps from single RGB images. Background pixels (higher depth values) are identified using quantile-based thresholding and removed:
foreground_mask = depth_map <= threshold
💡 Dynamic Brightness Adjustment
Adaptive image enhancement compensates for variable lighting in HSV color space:
brightness_factor = max_factor
elif avg_brightness < max_threshold:
brightness_factor = interpolate(avg_brightness)
🎯 YOLO Instance Segmentation
Custom-trained YOLO model with pixel-level flower segmentation generating polygon masks. Optimized with low confidence threshold (0.0001) to capture faint/partial flowers, IOU threshold (0.45) balancing false positives vs. negatives, and 640×640 input resolution for detail and efficiency.
📊 Spatial Clustering (DBSCAN)
Detected flowers grouped into biologically meaningful clusters with dynamically calculated epsilon:
clustering = DBSCAN(eps=epsilon, min_samples=1)
Iterates through epsilon factors (0.15–0.30) to achieve target cluster counts (~8 per image) matching typical branch structure.
🔢 Multi-Factor Density Calculation
Density percentage combines two independent metrics with configurable weights:
- •Flower Pixel %: Ratio of flower tissue pixels (white/light colors, multi-color-space thresholds: RGB, HSV, LAB) within cluster boundary
- •Edge Pixel %: Ratio of Canny edge pixels within flower pixels, indicating flower structure and density
Implementation Highlights
Depth-Anything-V2 Integration
Supports multiple ViT encoder sizes (Small/Base/Large/Giant). ViT-Large (256 features) default configuration. Pre-trained checkpoints with gradient-disabled evaluation mode for efficient inference.
YOLO Mask Polygon Extraction
Direct access to mask.xy coordinates preserving precise boundary information. Coordinates scaled from 640×640 inference to original image dimensions maintaining geometric accuracy.
Robust Polygon Geometry Handling
Comprehensive error handling for invalid geometries using Shapely's buffer(0) operation. Try-except blocks prevent pipeline failures from self-intersections and edge cases.
Multi-Color Space Flower Detection
Fuses RGB, HSV, LAB, and grayscale representations. Masks combined via bitwise OR creating comprehensive flower pixel classifier robust to color variation.
Batch Processing Pipeline
End-to-end orchestration: depth → brightness → YOLO → clustering → density. Generates individual and aggregated Excel reports with tqdm progress bars.
Visual Annotation System
Green polygon boundaries for clusters, red overlay (30% transparency) for flower pixels, green overlay (20%) for edge pixels, numbered labels with density percentages.
Technical Challenges & Solutions
⚡ Dense Flower Occlusions
✓ Instance segmentation with polygon masks captures overlapping flowers. Clustering + union operations merge partially occluded flowers into coherent representations without individual delineation
⚡ Invalid Polygon Geometries
✓ Shapely's buffer(0) repair operation and try-except blocks gracefully handle self-intersections, zero-area slivers, and geometric edge cases
⚡ Background Noise & False Positives
✓ Depth-based foreground segmentation removes background elements using geometric priors. Quantile-based threshold adapts to each image's depth distribution
⚡ Variable Lighting Conditions
✓ Dynamic brightness adjustment in HSV color space with multi-color-space flower detection (RGB/HSV/LAB/grayscale) ensures illumination robustness
⚡ Computational Efficiency
✓ CUDA GPU acceleration with CPU fallback, 640×640 YOLO inference resolution, NumPy vectorization, and batch processing amortizing model loading overhead
⚡ Cluster Size Variability
✓ Maximum area constraints (15-30% of image) prevent oversized clusters. Adaptive DBSCAN epsilon tuning with ~8 cluster target ensures biologically meaningful groupings
Applications & Impact
Yield Prediction
Early-season flower density estimates predict harvest volume, enabling better market planning and resource allocation
Crop-Load Management
Quantitative bloom density data informs thinning decisions (chemical/mechanical) to optimize fruit size and quality
Orchard Monitoring
Temporal density tracking across seasons identifies high/low-performing tree sections for targeted interventions
Research & Breeding
Automated phenotyping accelerates breeding programs by efficiently quantifying flowering traits across cultivars
Autonomous Systems
Pipeline integrates with UAVs, ground robots, or autonomous tractors for large-scale automated orchard assessment
Decision Support
Data-driven insights empower orchard managers with precision agriculture tools for sustainable production
Results & Validation
Future Enhancements
Project Impact
This Automated Flower Density Estimation System demonstrates the power of integrating modern computer vision techniques—monocular depth estimation, YOLO instance segmentation, spatial clustering, and multi-factor analysis—to solve complex agricultural challenges. By providing accurate, automated bloom density quantification, the system empowers orchard managers with data-driven insights for yield prediction, crop-load management, and precision thinning practices. The modular, configurable pipeline architecture ensures adaptability across diverse fruit crops and deployment scenarios, positioning it as a valuable tool for advancing precision agriculture.
Interested in This Project?
This project showcases advanced agricultural computer vision, deep learning deployment, and production-grade system development. Feel free to reach out for technical discussions or collaboration opportunities.